December 2020 Galactic News
And what a month it is
- **[Event news](#event-news)**:
- Webinars:
- Use Galaxy anywhere, right now, *December 9*
- Chloroplast Genome Assembly, *December 10*
- First Metabolomics Community Call, *December 10*
- Galaxy Developer Round Table: Working Groups, *December 10*
- Papercuts CollaborationFest: *December 17*
- Galaxy Admin Training: *Apply by December 18*
- Tripal CodeFest 2021: *January 11-15*
- GTN Smörgåsbord: A Global Galaxy Course. *Register by February 1*
- Webinars:
- **[Galaxy platform news](#galaxy-platforms-news):**
- CLIP-Explorer, a new server for CLIP-Seq data
- GenAP expansion funded by CANARIE (Canada!)
- UseGalaxy.eu gets more storage and GPUs through deNBI-Cloud
- Plus more UseGalaxy.\* news
- **[Blog posts](#galactic-blog-activity):**
- Outcome of the BioHackathon Europe
- Accessible single-cell RNA-sequencing bioinformatics training using Galaxy
- TIaaS and training feedback
- **[Training material and doc updates](#doc-hub-and-training-updates)**:
- Chloroplast genome assembly
- Compute and analyze Essential Biodiversity Variables with PAMPA toolsuite
- 16S Microbial analysis with Nanopore data
- **[Publications](#publications)**
- Metaproteomics, CLIP-Explorer, GIANT, and package management is your friend
- **[Q: Who's hiring?](#whos-hiring)**
- Europe:
- Max Planck IIE, University of Oslo, Hannover Medical School, EMBL Rome, Norwegian University of Life Science, Sorbonne-Université
- North America:
- Roche, Johns Hopkins (AnVIL, 2 positions; Galaxy, 2 Positions), Cleveland Clinic
- *A: Everyone, it seems.*
- Europe:
- **[New releases](#releases)**:
- Galaxy 20.09
- ENA submission
- OMERO accessible from Galaxy
- CloudLaunch
- **[Other news too](#other-news)**
If you have anything to include to next month's newsletter, then please send it to outreach@galaxyproject.org.
Event News
Despite COVID-19, there is still a lot going on. Some of it is virtual, but live events are starting to happen again, especially in Europe. We have updated our list of events to reflect what we know. Some highlights:

9 & 10 December, Online
Interested in learning more about Galaxy in a webinar? Here's two offered this month:
Use Galaxy on the web, the cloud, and your laptop too
There are many ways that researchers can access Galaxy. This webinar on Wednesday, December 9, will introduce participants to a range of options for using Galaxy instances around the world, and on your own laptop too.
Plant genomics: chloroplast genome assembly using Galaxy Australia
You can also see Galaxy, in action, in another webinar 12 hours later: Anna Syme of Australian BioCommons will present a webinar on plant genomics and genome assembly.

10 December, Online
The first call of the Galaxy Metabolomics Community will be held on the 10th of December at 2 PM CET. The bi-monthly community calls aim to bring together Galaxy metabolomics researchers -including users and developers- to discuss needs, ideas, and scientific problems, and to find new collaborators and synergies in the community. See the announcement for details.

10 December, Online
There will be one roundtable meetup this month. Discussion will focus on restructuring how development and other Galaxy components are gathered and coordinated.
A gentle shout form the editors: Wake up! If you want to get involved in guiding and contributing to Galaxy in a significant way, then please join this call.

17 December, Online
Paper Cuts are annoying but easy to fix bugs. October and November were a success, so we are going to do it again on December 17. Our third one-day Paper Cuts contribution fest will also be a 24-hour event spanning all time zones with our worldwide community.
Please save the date! It's an ideal opportunity for newcomers to become a Galaxy contributor.

Application Deadline: 18 December
Applications to attend the 2021 Galaxy Admin Training in January are now open and being accepted through 18 December. This week-long workshop will be online, global, and free. Apply now. (Applications are competitive).

11-15 January, Online
Calling all Tripal Core, Extension Module and Tool Integration Developers!
Tripal is a toolkit for construction of online biological (genetics, genomics, breeding, etc.), community database web portals. Tripal includes Tripal Galaxy and blend4php.

15-19 February, Online; Register by 1 February
This week-long workshop on how to use Galaxy will be online, global, and free. The program covers a general introduction to the Galaxy platform, NGS Analysis (DNA-seq and RNA-seq), Proteomics, and also features a Choose your own adventure day (!?).
Galaxy Platforms News
The Galaxy Platform Directory lists resources for easily running your analysis on Galaxy, including publicly available servers, cloud services, and containers and VMs that run Galaxy. Here's the recent platform news we know about:

The CLIP-Explorer server is a webserver hosted by the UseGalaxy.eu team to process, analyse and visualise CLIP-Seq data.
GenAP, a Galaxy provider for Canadian researchers has been awarded additional funds by CANARIE to expand it's service. Read more here.

The de.NBI-Cloud has extended the support to the European Galaxy Server by providing new computational infrastructure.
If you have some special needs that we should consider in our next purchase, let us know! And please consider growing the Galaxy computational resources with the funding leftovers of your group by the end of the year.


- Wanna know what has happened in the last 5 years in the Freiburg Galaxy Team? Here is the 5-years report!
- Lots of tool updates on UseGalaxy.eu and UseGalaxy.org.au.
Note: these platforms will feature prominently in the December 9 webinar.
Galactic Blog Activity
The Galaxy community participated in 8 different projects during the BioHackathon Europe 2020:
- CNV detection software containerisation and benchmark
- EDAM and Tool Information Profiles
- Exporting rich metadata and provenance from Galaxy using RO-Crate packaging
- Improve the support of Common Workflow Language in Galaxy
- Hardening and Testing Galaxy cluster support with BioContainer
- An interface between Galaxy and disease maps
- Deploying biocontainers in orchestration environments for life science research
- bio.tools integration and sustainable development

By Peter Bickerton.
“I wanted to step away from the command line”, Graham Etherington explains. “It takes a long time and it causes confusion, whereas everybody knows how to use a web browser."

By Ambre Jousselin.
Ambre reports on the Université de Toulouse Paul Sabatier's experience teaching Genomics using UseGalaxy.eu's Training Infrastructure as a Service (TIaaS).
By Anne Fouilloux, Research Software Engineer at NeIC.
This training covered modelling with FATES (Functionally Assembled Terrestrial Ecosystem Simulator) for improving climate models using UseGalaxy.eu's Training Infrastructure as a Service (TIaaS) and funded by EOSC-Life.

Doc, Hub, and Training Updates
By Coline Royaux and Yvan Le Bras.
This tutorial aims to present the PAMPA Galaxy workflow, how to use it to compute Essential Biodiversity Variables (EBV) from species abundance data and analyse it through generalized linear (mixed) models (GLM and GLMM). This workflow made up of 5 tools will allow you to process temporal series data that include at least year, location and species sampled along with abundance value and, finally, generate article-ready data products.

By Anna Syme.
This tutorial shows genome assembly for the plant chloroplast genome with a subset of a real data set from sweet potato.
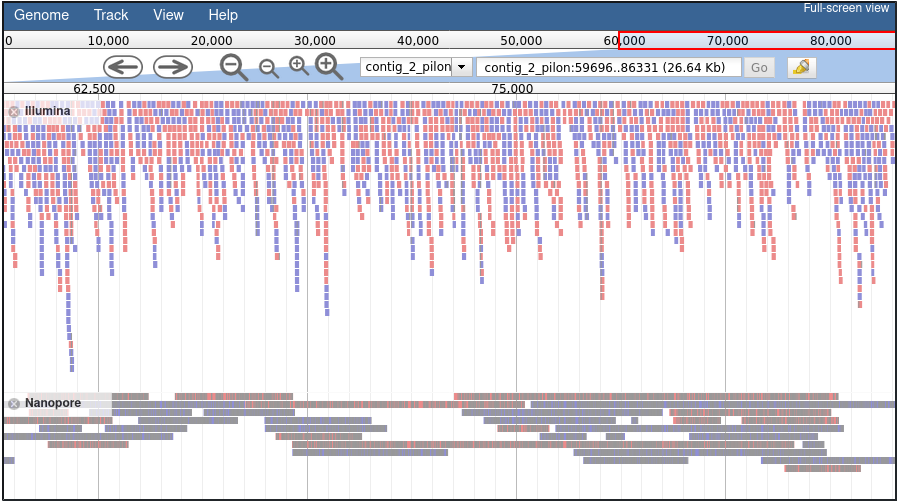
This tutorial uses sequencing data obtained through the MinION sequencer (Oxford Nanopore Technologies) with two objectives:
- evaluate the health status of soil samples,
- study how microbial populations are modified by their interaction with plant roots.

Publications
Pub curation activities are on hiatus right now but a few publications referencing, using, extending, and implementing Galaxy were added to the Galaxy Publication Library anyway. Here are the new open access Galactic and Stellar pubs:
Sajulga, R., Easterly, C., Riffle, M., Mesuere, B., Muth, T., Mehta, S., Kumar, P., Johnson, J., Gruening, B. A., Schiebenhoefer, H., Kolmeder, C. A., Fuchs, S., Nunn, B. L., Rudney, J., Griffin, T. J., & Jagtap, P. D. (2020). PLOS ONE, 15(11), e0241503. doi: 10.1371/journal.pone.0241503
Heyl, F., Maticzka, D., Uhl, M., & Backofen, R. (2020). GigaScience, 9(11). doi: 10.1093/gigascience/giaa108
Vandel, J., Gheeraert, C., Staels, B., Eeckhoute, J., Lefebvre, P., & Dubois-Chevalier, J. (2020). Scientific Reports, 10(1), 19835. doi: 10.1038/s41598-020-76769-w
Jalili, V., Clements, D., Gruning, B., Blankenberg, D., & Goecks, J. (2020). BioRxiv, 2020.11.16.385211. doi: 10.1101/2020.11.16.385211
Who's Hiring
Bioinformatics Unit, Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany
Work in an interdisciplinary team with close links to different departments, research groups, and other core facilities.
Apply by 11 December.
Centre for Bioinformatics (SBI), Department of Informatics, University of Oslo (UiO), Oslo, Norway.
This 3-year postdoc position will collaborate closely with the Oslo bioinformatics ELIXIR team, which is the Oslo node of the ELIXIR Norway project. Work on Genomic HyperBrowser, GTrack, FAIRtracks, TrackFind, Galaxy ProTo, and the Norwegian Federated European EGA (European Genome-Phenome Archive) node.
Apply by 11 December.

Research Core Unit Genomics (RCUG), Hannover Medical School, Hannover, Germany
A full position is available for 2 years in the Research Core Unit Genomics (RCUG), Germany, starting at the earliest timepoint possible. This includes working with and running training on the internal Galaxy instance.
Apply by 16 December.

Boskkovic Lab, EMBL Rome, Italy.
Directly participate in various experiments in developmental biology and epigenetics to meet the research goals of the laboratory.
Apply by 27 December.

MEMO Group, Norwegian University of Life Science, Ås, Norway
Interested in host-microbiome interactions and multi-omic data? We have multiple positions starting in 2021. Projects have fun and interesting EU partners. Will be hiring after Christmas.

Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University.
Data science research and education focusing on genomics (AnVIL, Genomic Data Science Community Network), cancer (ITCR) or pain A2CPS.

Roche, Bay Area, California, United States.
- Lead data mining for biomarker discovery for medical conditions of interest.
- Develop Agile Assay Design (AAD) tools for qPCR tests.
- NGS data analysis tools and/or workflows.
- Use these tools & workflows for R\&D projects.
- Deploy these tools on Roche intranet (Galaxy) and train scientists to use them.

Johns Hopkins University, Baltimore, Maryland, United States.
Provide technical expertise and oversight for the AnVIL Project, which incorporates Galaxy, Bioconductor, Terra, Gen3, and Dockstore into a secure cloud-based software ecosystem for genomic data analysis.

The Blankenberg Lab in the Genomic Medicine Institute at the Cleveland Clinic Lerner Research Institute is searching for Software Engineers and Postdoctoral Fellows. We utilize high-throughput omics technologies, such as Next-Generation Sequencing, and data-intensive computing to explore biomedical research questions.

A 3-year position of genomic data analyst is available to work within the "COllaborative NEtwork on research for Children and Teenagers with Acute Myeloblastic Leukemia" CONECT-AML framework.
The project is funded by the Institut National de Recherche sur le Cancer (INCA) and the Fight Kids Cancer programme. It involves 10 participant teams across France, with clinical or fundamental approaches.

The Schatz Lab at Johns Hopkins University is looking for:
- Self-driven individuals that can work independently to fill multiple software development positions on the Galaxy Project.
- Ambitious individuals to fill a programmer analyst position working on the Galaxy and AnVIL projects.

Releases
See
Features:
- GTN in Galaxy.
- Plugin framework for uploading datasets.
- Upload directly from the Tool Form.
- Workflow import from GA4GH TRS servers.
- Simplified workflow submission form.
- Accelerated batch job creation and workflow step scheduling.

ELIXIR Belgium, in collaboration with the European Galaxy project (de.NBI) and the European COVID-19 Data Platform, have developed a tool to simplify the submission of viral sequencing data to the European Nucleotide Archive (ENA), an ELIXIR Core Data Resource providing open access to nucleotide sequences. The new submission tool offers an easy-to-use interface, guides researchers through the submission process and verifies the data format and description. Read more in the official ELIXIR press release.
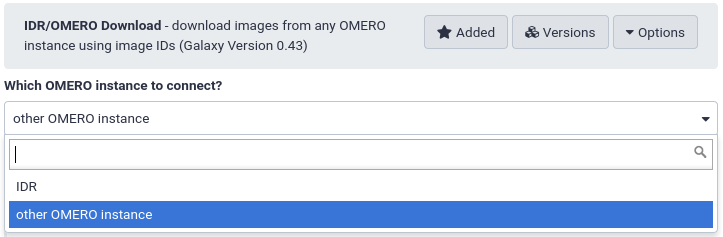
Good news for the imaging community! The IDR Download tool can now download images from a local OMERO instance.
This is an important step towards the accessibility of bioimage data. Now, images stored in local OMERO databases that are not publicly available are accessible from either public or private Galaxy instances. Imaging facilities that are running their local instance and want to perform image analysis in Galaxy can directly benefit from this new feature.
CloudBridge version 2.1.0 is a Python library that provides a consistent layer of abstraction over different Infrastructure-as-a-Service cloud providers, reducing or eliminating the need to write conditional code for each cloud. 2.1.0 adds new services and improved robustness. See the full documentation for more.
Other news
The Chan Zuckerberg Initiative's (CZI) Essential Open Source Software for Science (EOSS) program is funding work to extend Galaxy. See the announcement for details.


The JXTX Foundation sponsored 10 graduate students to attend the 2020 Biological Data Science Conference at Cold Spring Harbor Laboratory. The scholarships included connecting the recipients with prominent mentors in the field. Here what the awardees have to say about this experience: