Datatypes
Galaxy is designed to work with many different datatypes. Upon file upload, datatype can be detected and assigned (when possible) or user specified (before or after load). Datatype is also assigned by tools when output is created. It is important to note that many tools will only accept as input datasets with the appropriate datatype assigned.
A datatype can be altered in two ways:
- By the appropriate tool to transform the data from one type to another. See tool groups: Convert Formats, SAM Tools, and NGS: QC and manipulation, or perform a search in the top left Tool Search box.
- With the Edit Attributes form, reached by clicking on the pencil icon
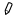 inside of a
dataset box in the history.
inside of a
dataset box in the history.
Tool developers: please see admin/datatypes for instructions about adding new datatypes to Galaxy.
Galaxy User Interface: "Format Help" accessed through datatype links on tool forms.
Ab1
A binary sequence file in ab1 format with a '.ab1' file extension. You must
manually select this datatype when uploading the file.
Axt
blastz pairwise alignment format. Each alignment block in an '.axt' file
contains three lines: a summary line and 2 sequence lines. Blocks are
separated from one another by blank lines. The summary line contains
chromosomal position and size information about the alignment. It consists of 9
required fields.
BAM
A binary file compressed in the BGZF format with a '.bam' file extension.
Also see SAM.
http://samtools.sourceforge.net/samtools.shtml
Bed
Tab delimited format (tabular) with a '.bed' file extension. Sometimes the
number of fields is noted in the file extension, for example: '.bed3',
'.bed4', '.bed6', '.bed12'. Valid BED files contain columns 1-3, 1-4,
1-5, 1-6 or 1-12.
Example BED12:
chr22 1000 5000 cloneA 960 + 1000 5000 0 2 567,488,0,3512
chr22 2000 6000 cloneB 900 - 2000 6000 0 2 433,399, 0,3601
Coordinates have a "0-based, fully-closed start" and a "0-based, half-open end"
and are reported with respect to the forward (+) strand. This is true for any
coordinate field in a BED file, including features aligned to the reverse (-)
strand. Simply put: the smallest coordinate is used as the "start" field, and
the largest coordinate is used as the "end" field (for a sequence aligned to
the reverse (-) strand, the "end" would be the beginning of the alignment and
the "start" would be the end). This can be confusing when first used.
Fortunately, an excellent discussion of the BED coordinate system (also used in
Interval and certain other
data formats) is at UCSC's genomewiki:
http://genomewiki.ucsc.edu/index.php/Coordinate_Transforms
For example, the first 100 bases of a chromosome would be given the coordinates: chromStart=0 and chromEnd=100.
- In a display application, a feature with these coordinates would overlap chromosome bases 1-100. This overlap could be on either the (+) or (-) strand, depending on the strand assignment in column 6.
- In a computational algorithm, a feature with these coordinates would
represent a chromosome base range with the mathematical notation of
[0,100), representing the 0-based numbered chromosome positions 0-99.
Also, see Interval format for more discussion about ""0-based, half-open start, fully closed end" coordinate system.
BED format does not require a header or custom track and/or browser line, but
may have one, starting with a "#" or "track" or "browser" or all three. If
extracted from UCSC's Table browser or Downloads area, a BED file may start
with a 'bin' column. This is an index number and the column should be removed
as a first step prior to any analysis (use tool Text Manipulation: Cut
columns). This Cut tool can also be used to rearrange the columns of an
Interval file to create a
BED file.
3 required fields:
| 1 | chrom | The name of the chromosome (e.g. chr3, chrY, chr2_random) or contig (e.g. ctgY1). |
| 2 | chromStart | The starting chromosome position of the feature. The start is 0-based, fully-closed. For example, the first base in a chromosome is numbered 0. |
| 3 | chromEnd | The ending chromosome position of the feature. The end is 0-based, half-open. |
9 optional fields:
| 4 | name | Defines the name of the BED line. A null or undefined name can be assigned as "." (a single dot, no quotes). |
| 5 | score | A score between 0 and 1000. A null or undefined score can be assigned as "0". |
| 6 | strand | Defines the strand - either '+' or '-'. A null or undefined strand can be assigned as "." (a single dot, no quotes). If strand is set to a "." or is absent, "+" strand is assumed. |
| 7 | thickStart | The starting position for the coding region (start codon) or other internal feature. |
| 8 | thickEnd | The ending position for the coding region (stop codon) or other internal feature. |
| 9 | itemRgb | An RGB value of the form R,G,B (e.g. 255,0,0). Is used for display in certain browsers. A null or undefined itemRgb value can be assigned as "0". |
| 10 | blockCount | The number of blocks (exons) in the BED line. |
| 11 | blockSizes | A comma-separated list of the block sizes. The number of items in this list should correspond to blockCount. |
| 12 | blockStarts | A comma-separated list of block starts. All of the blockStart positions should be calculated relative to chromStart. The number of items in this list should correspond to blockCount. |
The core field definitions above are based on the UCSC Genome Browser BED specification: http://genome.ucsc.edu/FAQ/FAQformat.html#format1
BedGraph
A tabular dataset that is line oriented and represents continuous-valued data. The format was developed at UCSC and the complete specification can be found here: http://genome.ucsc.edu/goldenPath/help/bedgraph.html.
BCF
Binary call format or "binary vcf" format has the extension .bcf. Also see
Variant call format or .vcf.
http://vcftools.sourceforge.net/specs.html
http://www.1000genomes.org/wiki/analysis/variant-call-format/bcf-binary-vcf-version-2
Dbn
The Dot-Bracket Notation format (.dbn) is a commonly used format to describe
2D RNA structures, in particular as output for structure predictors that can't
predict pseudoknots. Every entry consists of three lines:
- The header line, to provide information about the sequence (e.g. name, free energy).
- The sequence line, to provide the sequence of which the structures will be described.
-
The structure (Dot-Bracket) line, describing the structure of the sequence.
- Two nucleotides that form a bond are indicated with encloding parenthesis and dots represent unbound nucleotides.
- Pseudoknots are indicated with { .. } or [ .. ].
Example of a Dbn file:
>Sequence1 information - small hairpin (dG=-10.3) ggggaaacccc ((((...)))) >Sequence2 information (dG=0.0) aaaaaaaaaaa ...........
http://ultrastudio.org/en/Dot-Bracket_Notation
http://rna.urmc.rochester.edu/Text/File_Formats.html#DotBracket
Galaxy Dbn (Dot-Bracket notation) rules:
-
The first non-empty line is a header line: no comment lines are allowed.
- A header line starts with a '
>' symbol and continues with 0 or multiple symbols until the line ends.
- A header line starts with a '
-
The second non-empty line is a sequence line.
- A sequence line may only include chars that match the Fasta format
(https://en.wikipedia.org/wiki/FASTA_format#Sequence_representation)
symbols for nucleotides:
ACGTURYKMSWBDHVN, and may thus not include whitespaces. - A sequence line has no prefix and no suffix.
- A sequence line is case insensitive.
- A sequence line may only include chars that match the Fasta format
(https://en.wikipedia.org/wiki/FASTA_format#Sequence_representation)
symbols for nucleotides:
-
The third non-empty line is a structure (Dot-Bracket) line and only describes the 2D structure of the sequence above it.
- A structure line must consist of the following chars: '
.{}[]()'. - A structure line must be of the same length as the sequence line, and each char represents the structure of the nucleotide above it.
- A structure line has no prefix and no suffix.
-
A nucleotide pairs with only 1 or 0 other nucleotides.
- In a structure line, the number of '
(' symbols equals the number of ')' symbols, the number of '[' symbols equals the number of ']' symbols and the number of '{' symbols equals the number of '}' symbols.
- In a structure line, the number of '
- A structure line must consist of the following chars: '
- The format accepts multiple entries per file, given that each entry is provided as three lines: the header, sequence and structure line.
- Empty lines are allowed.
Fasta
A sequence in FASTA format consists of a single title line and one or more
lines of sequence data wrapped to a consistent length. The first character of
the title line is a greater-than (">") symbol.
>sequence1 atgcgtttgcgtgcatgcgtttgcgtgcatgcgtttgcgtgcatgcgtttgcgtgc gtcggtttcgttgcatgcgtttgcgtgcatgcgtttgcgtgcatgcgtttgcgtgc atgcgtttgcgtgc >sequence2 tttcgtgcgtatagtttcgtgcgtatagtttcgtgcgtatagtttcgtgcgtatag tttcgtgcgtatagtttcgtgcgtatagtttcgtgcgtatagtttcgtgcgtatag tggcgcggt
Galaxy FASTA format rules:
- the identifier name for a sequence is the first "word" after the ">" symbol in the title line
- no spaces are between the ">" symbol and the identifier name
- any description content in the title line after the first white space (space, tab, etc.) is ignored by most tools
- identifier names must be unique for each sequence contained within any
single
FASTAdataset - all sequence lines are wrapped to the same length (40-80 characters is recommended)
Best practices:
- confirm the format of a
FASTAdataset at the start of an analysis project - if a
FASTAdataset is used as a Custom Reference Genome, double check the formatting and fix it as needed. The tool NormalizeFasta can be used in most cases -- see the Custom Reference Genome FAQ for the how-to - use the same Target Fasta/Custom Genome dataset for all steps to avoid technical processing errors
Troubleshooting:
- Some tools will compensate for inconsistent sequence line wrapping, but
others will not. If a fasta dataset has inconsistent wrapping or is not
wrapped, use the tool
FASTA manipulation -> FASTA Width formatterto reformat. - Including blank lines in a
FASTAdataset will cause errors with many tools. Use the toolFilter and Sort -> Select lines that match an expressionwith the options "that: NOT Matching" and "the pattern: ^$" to remove. - Certain sources produce
FASTAdatasets with non-unique identifier names. Use the toolNGS: QC and manipulation -> Rename sequencesto assign unique identifier names.
For more details, please see the Wikipedia FASTA_format entry.
Fastq
While the basic FASTQ format remains somewhat constant, identifiers can
change quickly when new technologies are introduced. One of the best resource
for understanding current formats is the Wikipedia
FASTQ_format entry.
When using Galaxy, many tools require that input FASTQ files be standardized
as the first step in an analysis. Use the tool NGS: QC and manipulation -> FASTQ Groomer. Instructions on the tool form explain how to use and the
resulting output format.
FastqSolexa
FastqSolexa is the Illumina (Solexa) variant of the Fastq format, which
stores sequences and quality scores in a single file:
@seq1
GACAGCTTGGTTTTTAGTGAGTTGTTCCTTTCTTT
+seq1
hhhhhhhhhhhhhhhhhhhhhhhhhhPW@hhhhhh
@seq2
GCAATGACGGCAGCAATAAACTCAACAGGTGCTGG
+seq2
hhhhhhhhhhhhhhYhhahhhhWhAhFhSIJGChO
Or:
@seq1
GAATTGATCAGGACATAGGACAACTGTAGGCACCAT
+seq1
40 40 40 40 35 40 40 40 25 40 40 26 40 9 33 11 40 35 17 40 40 33 40 7 9 15 3 22 15 30 11 17 9 4 9 4
@seq2
GAGTTCTCGTCGCCTGTAGGCACCATCAATCGTATG
+seq2
40 15 40 17 6 36 40 40 40 25 40 9 35 33 40 14 14 18 15 17 19 28 31 4 24 18 27 14 15 18 2 8 12 8 11 9
GFF
GFF lines have nine required fields that must be tab-separated.
- Fields are:
- seqname
- source
- feature
- start (1-based)
- end
- score
- strand
- frame
- group
GFF format is also known as General Feature Format 1 or GFF1. The official specification is at
http://www.sanger.ac.uk/resources/software/gff/spec.html
(notes in the GFF2 specification describe
GFF1).
The UCSC Genome Browser GFF
specification: http://genome.ucsc.edu/FAQ/FAQformat.html#format3
GTF
GTF lines have nine required fields that must be tab-separated. (Similar to
GFF format)
- Fields are:
- seqname
- source
- feature
- start (1-based)
- end
- score
- strand
- frame
- attribute
GTF format is also known as General Feature Format 2 or GFF2. The official specification is at
http://www.sanger.ac.uk/resources/software/gff/spec.html
(GFF2 specification).
The UCSC Genome Browser GTF
specification: http://genome.ucsc.edu/FAQ/FAQformat.html#format4
TIP When using GTF datasets in Galaxy, the dataset should not contain any header lines or blank lines. Extra comment lines (usually staring with a #) or blank lines at the start and sometimes internal to the file should be removed before using the data.
- Remove the headers (lines that start with a "#") with the Select tool using the option "NOT Matching" with the regular expression:
^# - Remove blank lines with the Select tool using the option "NOT Matching" with the regular expression:
^$ - Once the formatting is fixed, change the datatype to be
gftunder Edit Attributes (pencil icon). - Often
gftdata will be given the datatypegffby default when formatting problems are present, which works fine with some tools and but not with others. It is a good idea to fix the data first, at the start of an analysis, to avoid confusing tool errors.
GFF3
Similar to GFF and GTF in
having nine tab-seperated columns of data at the core of file and having a
1-based start coordinate. However, GFF3 format has data that is grouped
differently between lines (and sets of lines), can be hierarchically ordered,
and can contain extra content such as FASTA sequence.
Seeing the official specification (and online validation tool) for details is
highly recommended.
TIP When using GFF3 datasets in Galaxy, the dataset must containly only the single header line and the primary data lines or tools may error. Extra comment lines (###), repeats, and fasta content is not accepted.
When obtaining reference annotation from the Ensembl downloads area, choose the
GTFannotation for use with Galaxy's tools. Avoid theGFF3annotation as it contains this extra content.
Known as General Feature Format
3, or GFF3.
The official specification is at http://www.sequenceontology.org/gff3.shtml.
GMOD also has a nice specification at GFF3_Format.
GFF/GTF/GFF3 datasets are available from many sources and can sometimes be created from other datatypes.
- How to source Human, Mouse, and other common genome
GTFreference annnotation data. See Method 6 here. - The public Main Galaxy instance contains tools to examine and manipulate GFF/GTF files under the tool group Filter and Sort.
- The public Galaxy instance at: http://galaxy.raetschlab.org contains tools to examine GFF files or convert various formats to GFF3 under the tool group GFF Toolkit.
- The Tool Shed contains a repository named
fml_gff3togtfthat houses a collection of tools to convert various formats into GFF3 format. - New tools are continually added to the Tool Shed - search with a phase like "GFF" to see if others are available.
Interval
Interval format, also known as "Genomic Intervals" is a datatype developed by the Galaxy team. This format incorporates a 0-based genome coordinates and labels in a tab-delimited file with no requirements on column order. (See BED format for a discussion of the ""0-based, half-open start, fully closed end" coordinate system ). Or, this quick reference:
0-relative positions, half-open start, closed stop: Interval, BED, others [0,100) == 0-100 in files == 0-99 in calculations == 1-100 in display
1-relative positions, closed start and closed stop: MAF, others [1,100] == 1-100 in files == 1-100 in calculations == 1-100 in display
Interval dataset start with definition line that assigns the column
attributes. Columns are individually assigned to an Interval attribute
CHROM, START, END, NAME, STRAND, SCORE, COMMENT. The columns may be in any
order and only CHROM, START, and END are required. To add/assign additional
columns, use the "Edit Attributes" form (click on pencil icon in top right
corner of dataset).
Most common bioinformatics coordinate data formats can be interpreted as or
transformed to Interval format. For example: '.bed' datasets (always),
0-based coordinate '.tab' or '.txt' datasets, 1-based '.GFF/.GTF/.GFF3'
datasets (use Convert Formats -> GFF-to-BED), and others.
Example definition line:
#CHROM START END STRAND NAME SCORE COMMENT
| 1 | CHROM |
The name of the chromosome (e.g. chr3, chrY, chr2_random) or contig (e.g. ctgY1). |
| 2 | START |
The starting chromosome position of the feature. The first base in a chromosome is numbered 0. See BED format (above). |
| 3 | END |
The ending chromosome position of the feature. For example, the first 100 bases of a chromosome are defined as chromStart=0, chromEnd=100. |
| 4 | STRAND |
Defines the strand - either '+' or '-'. Optional value, null or undefined is omitted or a "." ("dot" without quotes). |
| 5 | NAME |
Name for the feature. Free text, no whitespace. Optional value. |
| 6 | SCORE |
Numeric value for the feature used for calculations and display. Optional value. |
| 7 | COMMENT |
Free text, no whitespace. Optional value. |
Example:#CHROM START END STRAND NAME COMMENT chr1 10 100 + exon myExon chrX 1000 10050 - gene myGene
Lav
Lav is the primary output format for BLASTZ. The first line of a '.lav'
file begins with '#:lav.'.
MAF
TBA and multiz multiple alignment format. The first line of a .maf file begins
with ##maf. This word is followed by white-space-separated "variable=value"
pairs. There should be no white space surrounding the =.
SAM
Also see BAM. http://samtools.sourceforge.net/samtools.shtml
Scf
A binary sequence file in 'scf' format with a '.scf' file extension. You
must manually select this 'File Format' when uploading the file.
Sff
A binary file in 'Standard Flowgram Format' with a '.sff' file extension.
Tabular (tab delimited)
Any data in tab delimited format (tabular). May end with the extension '.tab'
or '.txt'.
VCF
Variant call format has a few different active versions, depending on the
source. The first comment line will note which you have. These external links
can help with understanding the different .vcf types. It is important to note
that many tools will not interpret v4.1 content correctly and it should be
avoided as input (as of mid-2013). A compressed version of this data is the
Binary call format or "binary vcf" format .bcf.
Current VCF specification:
Alternate and Prior:
- http://vcftools.sourceforge.net/specs.html
- http://en.wikipedia.org/wiki/Variant_Call_Format
- http://www.1000genomes.org/wiki/Analysis/vcf4.0
- http://www.1000genomes.org/wiki/Analysis/Variant%20Call%20Format/vcf-variant-call-format-version-41
Wig and bigWig
The wiggle format is 1-based, line-oriented, and ends with the extension
'.wig'. Wiggle data is preceded by a required track definition line, which
adds a number of options for controlling the default display of this track. The
wiggle (WIG) format is for display of dense, continuous data. When large, using
the related .bigWig format is recommended.
This information and the .wig and .bigWig formats were developed at
UCSC. The official specifications are at:
- http://genome.ucsc.edu/goldenPath/help/wiggle.html
- https://genome.ucsc.edu/goldenPath/help/bigWig.html
Plain text
Any plain text file, commonly ending with the file extension ".txt".