COMBAT-TB Workbench
Launch Galaxy| Container: | COMBAT-TB Docker |
|---|---|
| Scope: | Domain Specific |
| Summary: | A modular, easy to install application that provides a web based environment for Mycobacterium tuberculosis bioinformatics |
Comments
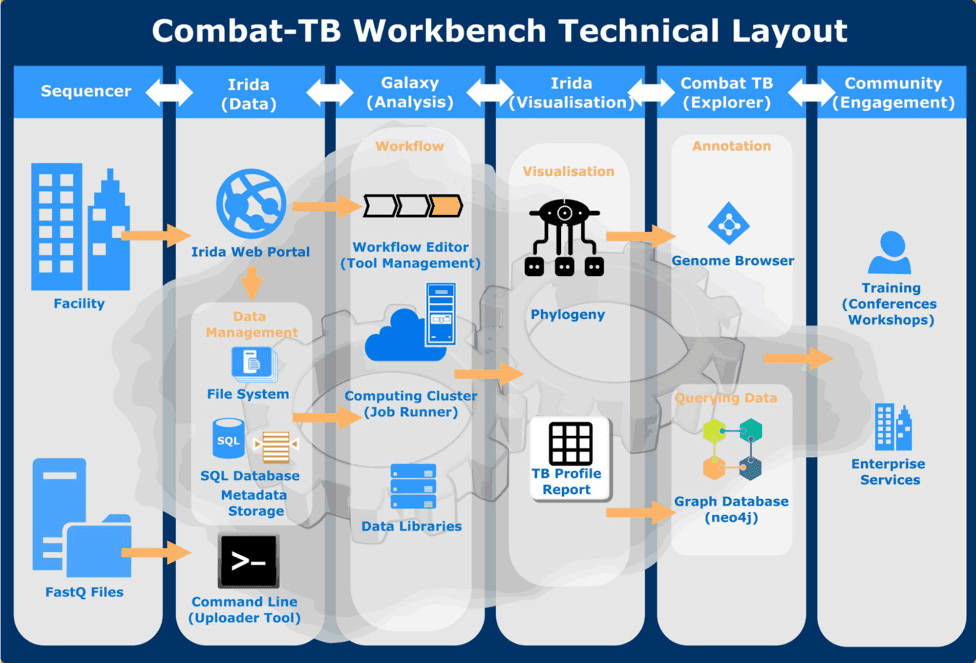
- COMBAT-TB Workbench is built using two main software components: the IRIDA Platform for its web-based user interface and data management capabilities and the Galaxy bioinformatics workflow platform for workflow execution.
User Support
Quotas
- None
Citations
- Heusden, P. van, Mashologu, Z., Lose, T., Warren, R., & Christoffels, A. (2021). The COMBAT-TB Workbench: Making powerful M. tuberculosis bioinformatics accessible. medRxiv, 2021.09.23.21263983. https://doi.org/10.1101/2021.09.23.21263983
- COMBAT-TB tagged publications in the Galaxy Publication Library
Sponsors
- South African Medical Research Council Bioinformatics Unit, South African National Bioinformatics Institute, University of the Western Cape, South Africa