May 2020 Galactic News
James Taylor, BCC2020, COVID-19 Response, and more
From the editor
This is our first newsletter since January. It has been an eventful and sorrowful four months for the world, and for the Galaxy Community too: This newsletter starts with the tragic loss of James Taylor, one of Galaxy’s founders and leaders. We lost James at the beginning of April. This community, I suspect, will always feel that loss.
This newsletter also covers how Galaxy is addressing the international COVID-19 crises, and how the pandemic pushed BCC2020 organizers to shift from an in-person event in Toronto, to a truly global, affordable, and accessible conference, where any researcher in the world can now participate. Even in the darkest of times, there is some sunlight.
The mix of news this month reflects our times. Our support of each other, no matter what, reflects the strength of this community.
Thanks for everything, and please continue to support each other,
Dave Clements, on behalf of the Galaxy Community
In the May 2020 issue
- James Peter Taylor, 1979-2020
- BCC2020 will be online, global, affordable, and accessible
- All abstracts are due May 8
- Galaxy COVID-19 Response
- Upcoming events
- Blog posts
- Galaxy Platform News
- Training material and doc updates
- Who’s Hiring
- New Releases
- New publications (671 of them)
- And other cool news too
If you have anything to include to next month’s newsletter, then please send it to outreach@galaxyproject.org.
James Peter Taylor, 1979-2020
James Taylor, one of the founders and leaders of the Galaxy Project died of natural causes on April 2. One day he was online tweeting about open access to data, and the next day he was not. News of his passing spread around the world, and the response has been overwhelming.
These responses, plus a summary of his academic life, and extended remembrances from several colleagues have been compiled on the @jxtx page. If you want to add your thoughts, please submit them here and we will post them.
We are also starting a foundation to continue and commemorate James’ work by supporting grad students, junior faculty, and underrepresented groups. Please consider contributing.
Galaxy will go on and we will continue to support his legacy of open reproducible science.
We miss you James.
BCC2020 will be Online, Global, Affordable, and Accessible

The 2020 Bioinformatics Community Conference (BCC2020) brings together the Bioinformatics Open Source Conference (BOSC) and Galaxy Community Conference. If you are working in data intensive life science research then there is no better event for sharing your work, and learning from other researchers addressing the challenges of modern data driven biology. BCC2020 will be held July 17-26, and offer 2 days of training, a 3 day meeting, and a 4 day CollaborationFest.

All BCC2020 events will be held online. Training will be live and interactive. The meeting will feature keynotes, accepted talks, lightning talks, posters, demos, and birds-of-a-feather and other networking opportunities. Talks (with the possible exception of keynotes) will be pre-recorded. Posters, demos, and BoFs will be live and interactive. The CoFest will also be live and interactive.
BCC2020 events will be held twice: once in the originally scheduled Toronto time zone (BCC West/Americas), and then again 12 hours later in the Eastern hemisphere (BCC East/Asia-Australia). Training will differ between East and West, with enrollment open to all, regardless of where you are. The main conference content will be presented in both East and West. We are striving to have the CoFest run continuously, with participants from every part of the world.

We have slashed registration rates for BCC2020, and are offering even larger discounts to participants based in low and lower-middle income countries. Pricing starts at US$3 per training session, and $12 for the 3 day meeting. The CoFest is free.

Going online and global, combined with the low registration rates this enables, makes this the most accessible Galaxy or BOSC conference ever. If you work in open source bioinformatics, anywhere in the world, then this is 2020’s best opportunity to share your work and learn from others.

We are pleased to announce that Abigail Cabunoc Mayes of the Mozilla Foundation, and Lincoln Stein of OICR will be keynote speakers at BCC2020.

BCC2020 is seeking oral presentations, lightning talks, posters, and demos, from researchers working in bioinformatics, and all over the world. Abstracts are due May 8 (and that deadline will not be extended). Please submit your work today.

BCC2020 registration is now open. Registering early saves 50% off of the full rates and starts $3 per training session and $12 for the three day meeting.
Galaxy COVID-19 Response
A wide variety of Galaxy community member organizations are contributing and collaborating to help address the coronavirus pandemic.
UseGalaxy.* COVID-19 Efforts
Several prominent efforts use entirely open source tools using open access data, on public cyberinfrastructure. Galaxy workflows and histories are provided by all analyses (in both Galaxy and Zenodo), making this work easily accessible and reusable by all. The work produced by this consortium is documented and runnable in the UseGalaxy.* servers, and available in Zenodo as well.
These efforts focus on three areas:

There are 397 sites showing intra-host variation across 33 samples (with frequencies between 5% and 95%). Twenty nine samples have fixed differences at 39 sites from the published reference. Variant lists and VCF files are updated daily.

We are using comparative evolutionary techniques to run daily analyses identify potential candidates using genomes from GISAID. At present, ~5 genomic positions may merit further investigation because they may be subject to diversifying positive selection. See live results presented as continuously updated notebooks.

Computational analyses using protein-ligand docking to identify potentially inhibitory compounds that can bind to MPro and can be used to control viral proliferation. This work analyzed over 40,000 compounds considered to be likely to bind, which were chosen based on recently published X-ray crystal structures, and identified 500 high scoring compounds.
Additional Efforts
And there are many additional efforts and posts about COVID-19 research using Galaxy:

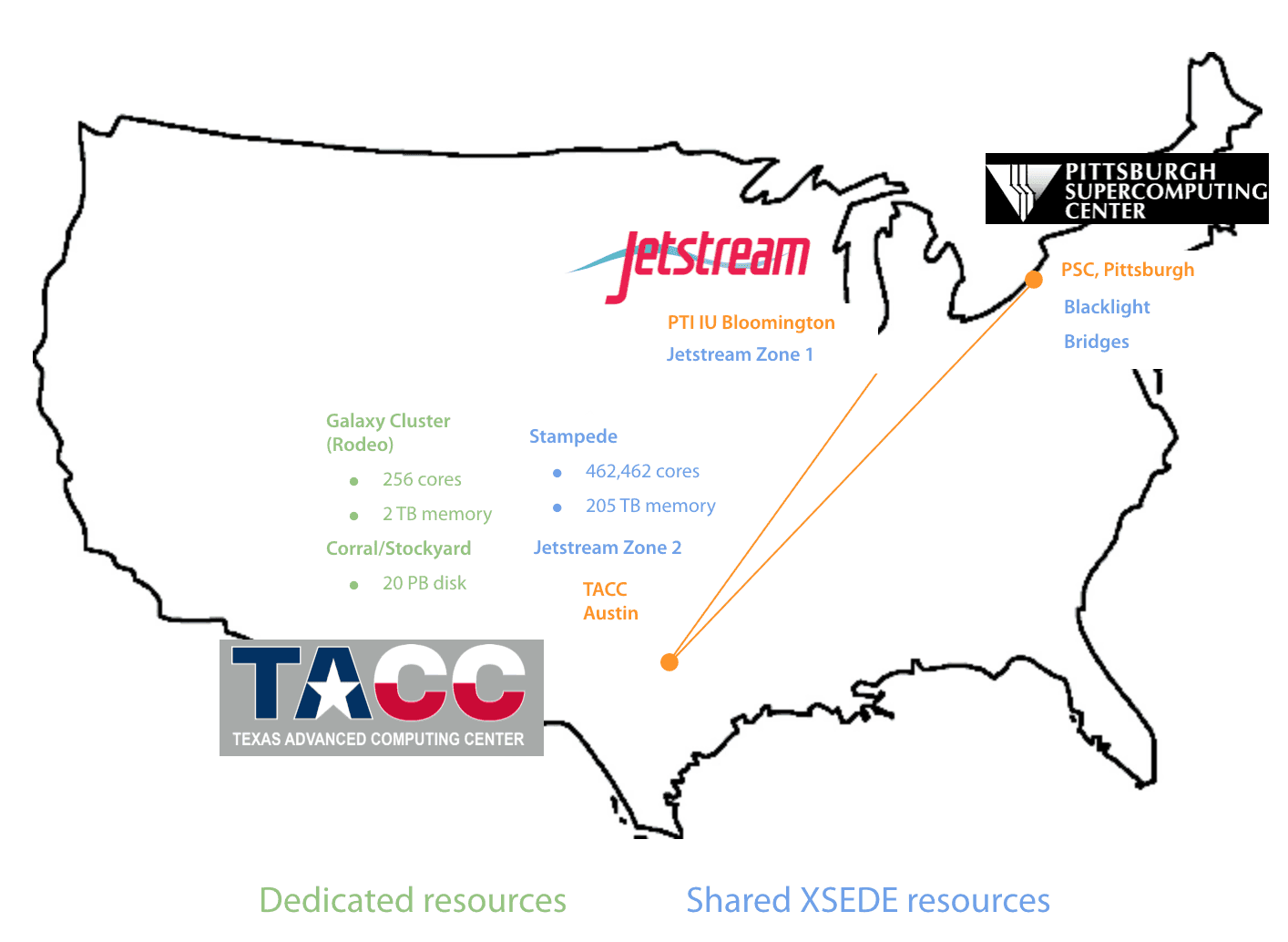
The Texas Advanced Computing Center (TACC) provides large-scale compute infrastructure for the analysis of thousands of genomes, including Galaxy’s work on SARS-CoV-2.

The MRC CLIMB project has been providing compute and storage for the COVID-19 Genomics UK Consortium (COG-UK). (See below for more CLIMB news.)

This new Galaxy Training Network tutorial from Simon Bray is a companion tutorial for the cheminformatics work described above that performs virtual screening on candidate ligands for the SARS-CoV-2 main protease (MPro).

This new Galaxy Training Network tutorial from Wolfgang Maier guides you through the preprocessing of sequencing data of bronchoalveolar lavage fluid (BALF) samples obtained from early COVID-19 patients in China. Since such samples are expected to be contaminated signficantly with human sequenced reads, the goal is to enrich the data for SARS-CoV-2 reads by identifying and discarding reads of human origin before trying to assemble the viral genome sequence.

A Galaxy Covid-19 flavour is now available in Laniakea, as a Docker Container. It is based on the GalaxyProject covid-19 analysis and it is continuously updated. Due to the current Covid-19 outbreak, the flavour is made available to Laniakea users without the usual test routine.

Galaxy Australia relies on distributed deployments using Pulsar to increase the range and number of jobs that can be run on the service. The team has been allocated resources on the Nimbus cloud to deploy a dedicated COVID-19 Pulsar as part of Galaxy Australia at the Pawsey Centre that allows Galaxy users to rapidly analyse their data on published tools/workflows to further research into SARS-CoV-2.
Upcoming Events
The coronavirus outbreak has impacted BCC2020, and just about every other event for the rest of the year too. Most events through the end of August have been postponed or moved online. We have updated our list of events to reflect what we know. Some highlights:

FAIR data and Open Infrastructures to tackle the COVID-19 pandemic
This webinar series demonstrates how open access and open science are fundamental for fast and efficient response to public health crises. The focus will be on research reproducibility and transparency, using exclusively open source tools and the Galaxy platform.
The first session was held on 30 April. Subsequent sessions are
- Genomics/Variant Calling, 7 May, Anton Nekrutenko and Wolfgang Maier
- Cheminformatics: Screening of the main protease, 14 May, TBA
- Evolution of the Virus, 20 May, Sergei Pond
- Behind the scenes: Global Open Infrastructures at work, 28 May, TBA
Want to learn the Galaxy community and platform big picture? Attend the next two Scientific Gateways Community Institute webinars:
- Galaxy Project: Enabling an active global research community, May 13, Dave Clements
- Overview of the Galaxy Project platform, June 10, Nate Coraor

There are
- 25 upcoming events (most of them virtual)
- covering COVID-19 (5 events), single-cell, variant detection, assembly, RNA-Seq and more.
And material from some recent past events is now available:
- Galaxy: much more than a workflow management system: Video
- Galaxy @ PAG 2020: Slides and posters
- Development of BioCompute Objects for Integration into Galaxy in a Cloud Computing Environment: on video
- Intro to Circos, its applications & use within Galaxy Australia: on video
- Galaxy-based Multi-omic Informatics Hub for Cancer Researchers: on video
Galactic Blog Activity
By Björn Grüning.
A visualization plug-in that extends Galaxy-P’s advantages into the visualization of large, complex datasets.

By Michael Thompson.
Michael Thompson of Kwame Nkrumah University of Science and Technology (K.N.U.S.T) describes his experience at the 2020 Galaxy Admin Training in Barcelona.

By Magnus Ø. Arntzen.
Adaption of a repertoire of commonly used omics tools spanning metagenomics, -transcriptomics and -proteomics into the Galaxy framework, in order to generate a user-accessible, scalable and robust analytical pipeline for integrated meta-omics analysis.

Galaxy Platforms News
The Galaxy Platform Directory lists resources for easily running your analysis on Galaxy, including publicly available servers, cloud services, and containers and VMs that run Galaxy. There are many new platforms this month:

The CLIMB project (Cloud Infrastructure for Microbial Bioinformatics) has been renewed as the CLIMB-BIG-DATA project. The initiative will benefit from a just-awarded £2Million grant from the UKRI, and will gradually become self-sustaining. This will ensure long-term provision of an always up-to-date cloud-based infrastructure for microbial bioinformatics.

The CoralSNP server implements Standard Tools for Acroporid Genotyping (STAG). In STAG the user’s data is compared to the database of previously genotyped samples and generates a report of genet identification. A login is required, but anyone can create a login.

The Mississippi server was upgraded, and has a new URL. Every tool installed on the previous server should be already installed on the new server. The old server will be put into a read only state on June 1st, and then taken down on September 1st.

The ELIXIR-ITALY Laniakea@ReCaS Call offers access to Cloud resources to be used for the deployment of on-demand Galaxy instances, ready for production, with reference data and tools already pre-configured and ready to be used.

ProteoRE 2.1 is a user-oriented Galaxy-based service for the functional interpretation and exploration of proteomics data for biomedical research; This version now comprises 20 tools organized in 4 sections (data manipulation and visualization; add features/annotation; functional analysis; pathways analysis). All data sources have been updated. Two tutorials are available via the Galaxy Training Network.

- Galaxy Australia processed its one millionth job in January.
- Exciting progress in the ELIXIR Galaxy community
- ChiRA, an integrated framework for Chimeric Read Analysis from RNA-RNA interactome data, added to RNA Workbench URL
- UseGalaxy.eu reaches 12,000 users, 6,900,000 jobs, and 13,300,000 datasets
Platforms that were referenced at least three times in recent publications:
103 : Huttenhower 24 : RepeatExplorer 20 : Workflow4Metabolomics 15 : UseGalaxy.eu 14 : ARGs-OAP 11 : CPT 8 : Galaxy-P 7 : UseGalaxy.org.au 6 : Cistrome 5 : Globus Genomics 5 : LAPPS Grid 4 : UseGalaxy.org 3 : deepTools 3 : GVL-Unspecified 3 : HiCExplorer 3 : Langille 3 : Pasteur 3 : PhenoMeNal 3 : RiboGalaxy
Doc, Hub, and Training Updates
The Galaxy Training Network library has been entirely updated to reflect current best practices and new features implemented in the last year. If you are learning Galaxy admin, this is where you should start.

By Melanie Foell and Matthias Fahrner.
Introduces the data analysis from raw data files to protein identification and quantification of two label-free human serum samples with the MaxQuant software.

Familiarze yourself with the Panoply Galaxy interactive environment. Panoply is among the most popular tools to visualize geo-referenced data stored in Network Common Data Form (netCDF).

We’ve seen the TIaaS Queue Status receive a lot of positive feedback. Helena Rasche has added two new features to get a general information about the TIaaS service.
- A calendar that shows when TIaaS trainings are booked
- Some Statistics about the TIaaS events

By Helena Rasche and Saskia Hiltemann.
How to set up your own Training Infrastructure as a Service (TIaaS) service to support Galaxy training compute infrastructure.

By Jennifer Hillman-Jackson.
- How to find hard-to-locate histories and datasets
- New guidelines for downloading large datasets and troubleshooting downloads
- Account quotas

By Nate Coraor.
Find out the latest about how the UseGalaxy.org server is set up.
OIDC is OpenID Connect, a simple identity layer on top of the OAuth 2.0 authorization protocol. Galaxy supports it.
- Configure Your Galaxy Instance as a CILogon OIDC Client, by Juleen Graham.
- Login to Galaxy Using Your Organization’s Okta identity, by Peter Selten
- Configure Your Galaxy Instance as an OIDC Client for your organization’s Okta Infrastructure, by Peter Selten

By Pavankumar Videm.
This GTN tutorial presents the analysis of a CLEAR-CLIP data set using the ChiRA tool suite.

By Anup Kumar and Alireza Khanteymoori
What are deep learning and neural networks? Why is it useful? How to create a neural network architecture for classification? This tutorial presents basic principles of deep learning.

By Alireza Khanteymoori, Anup Kumar and Simon Bray
How to use regression techniques to create predictive models from biological datasets.

By Pratik Jagtap, Subina Mehta, Ray Sajulga, Bérénice Batut, Emma Leith, Praveen Kumar, and Saskia Hiltemann.
This is a shortened version of an existing tutorial. Instead of running each tool individually, this tutorial employs workflows to run groups of analysis steps (e.g. data cleaning) at once.

Who’s Hiring
- Senior Software Developer, Black Canyon Consulting, Bethesda, Maryland, United States
- Bioinformatics Software Developer, AbSci, Vancouver, Washington, United States
- Data Scientist, New England Biolabs, Ipswich, Massachusetts, United States
Releases
See
Features:
- Easily list and review recently invoked workflows.
- Galaxy Markdown Pages and Workflow Reports as PDF
- Screenreader-friendly Navigation
- Email notification for completed jobs
- Workflows can now make use of optional datasets and optional parameters
- Major update to container and dependency management interface Extended job metadata collection

By Alexandru Mahmoud, Nuwan Goonasekera, Luke Sargent, Enis Afgan, Alex Ostrovsky, and the GVL and Galaxy teams.
GVL, the Genomics Virtual laboratory, had two beta releases in the first 4 months of 2020:
- With Love: The All-new GVL 5.0 (beta): Now more reliable, with better security, and with new features.
- GVL 5.0-beta2 release: 30% faster and single sign-on, ohh my
The GVL makes dedicated, production-grade installations of Galaxy available on cloud providers, all via a web browser. The GVL has been used extensively whenever public and shared servers were not suitable. The GVL 5.0 is a ground-up rewrite of the GVL based on Kubernetes and containerization technologies.
Galaxy Helm 3.1.0 was also released simultaneously.

Command-line utilities to help with managing users, data libraries and tools in a Galaxy instance, using the Galaxy API via the Bioblend library.

Galaxy’s sequence utilities are a set of Python modules for reading, analyzing, and converting sequence formats.
Publications
671 new publications referencing, using, extending, and implementing Galaxy were added to the Galaxy Publication Library in January, February, and March. There were over 25 Galactic and Stellar publications added, and 20 of them are open access:
Moreno, P., Huang, N., Manning, J. R., Mohammed, S., Solovyev, A., Polanski, K., Chazarra, R., Talavera-Lopez, C. A., Doyle, M., Marnier, G., Gruening, B. A., Rasche, H., Bacon, W., Perez-Riverol, Y., Haeussler, M., Meyer, K. B., Teichmann, S., & Papatheodorou, I. (2020). BioRxiv, 2020.04.08.032698. https://doi.org/10.1101/2020.04.08.032698
Werner, S., Schmidt, L., Marchand, V., Kemmer, T., Falschlunger, C., Sednev, M. V., Bec, G., Ennifar, E., Höbartner, C., Micura, R., Motorin, Y., Hildebrandt, A., & Helm, M. (2020). Nucleic Acids Research. https://doi.org/10.1093/nar/gkaa113
Sajulga, R., Easterly, C., Riffle, M., Mesuere, B., Muth, T., Mehta, S., Kumar, P., Johnson, J., Gruening, B., Schiebenhoefer, H., Kolmeder, C. A., Fuchs, S., Nunn, B. L., Rudney, J., Griffin, T. J., & Jagtap, P. D. (2020). BioRxiv, 2020.01.07.897561. https://doi.org/10.1101/2020.01.07.897561
Eisler, D., Fornika, D., Tindale, L. C., Chan, T., Sabaiduc, S., Hickman, R., Chambers, C., Krajden, M., Skowronski, D. M., Jassem, A., & Hsiao, W. (2020). Influenza and Other Respiratory Viruses. https://doi.org/10.1111/irv.12722
Miladi, M., Sokhoyan, E., Houwaart, T., Heyne, S., Costa, F., Grüning, B., & Backofen, R. (2019). GigaScience, 8(12). https://doi.org/10.1093/gigascience/giz150
Stoler, N., Arbeithuber, B., Povysil, G., Heinzl, M., Salazar, R., Makova, K. D., Tiemann-Boege, I., & Nekrutenko, A. (2020). BMC Bioinformatics, 21(1), 96. https://doi.org/10.1186/s12859-020-3419-8
Berrios, D., Weitz, E., Grigorev, K., Costes, S., Gebre, S., & Beheshti, A. (2020). EPiC Series in Computing, 70, 89–98. https://doi.org/10.29007/rh7n
Galaxy and HyPhy developments teams, Nekrutenko, A., & Pond, S. L. K. (2020). BioRxiv, 2020.02.21.959973. https://doi.org/10.1101/2020.02.21.959973
Defelicibus, A. (2016). [Thesis, Universidade de São Paulo]. https://doi.org/10.11606/D.82.2016.tde-22062016-102823
Chiara, M., Mandreoli, P., Tangaro, M. A., D’Erchia, A. M., Sorrentino, S., Forleo, C., Horner, D. S., Zambelli, F., & Pesole, G. (2020). BioRxiv, 2020.01.23.917229. https://doi.org/10.1101/2020.01.23.917229
Codó Tarraubella, L. (2019). [Thesis, Universitat de Barcelona]. http://diposit.ub.edu/dspace/handle/2445/149802
Holthausen Bermejo, R. (2019). [Thesis, Universidad de Malaga]. https://riuma.uma.es/xmlui/handle/10630/19060
El-Haj, M., Rutherford, N., Rayson, P., Knight, J., Piao, S., Coole, M., Mariani, J., Ezeani, I., Prentice, S., Ide, N., & Suderman, K. (2020, March 11). LREC 2020, Twelfth International Conference on Language Resources and Evaluation. https://eprints.lancs.ac.uk/id/eprint/142283/
Uellendahl-Werth, F., Wolfien, M., Franke, A., Wolkenhauer, O., & Ellinghaus, D. (2020). Scientific Reports, 10(1), 1–10. https://doi.org/10.1038/s41598-020-62637-0
Martin, M. (2020). [Thesis, UCL - Université Catholique de Louvain]. https://dial.uclouvain.be/pr/boreal/object/boreal:227671
Poncheewin, W., Hermes, G. D. A., van Dam, J. C. J., Koehorst, J. J., Smidt, H., & Schaap, P. J. (2020). Frontiers in Genetics, 10. https://doi.org/10.3389/fgene.2019.01366
Hernández, I., Sant, C., Martínez, R., & Fernández, C. (2020). Frontiers in Microbiology, 11. https://doi.org/10.3389/fmicb.2020.00208
Kitchen, S. A., Kuster, G. V., Kuntz, K. L. V., Reich, H. G., Miller, W., Griffin, S., Fogarty, N. D., & Baums, I. B. (2020). BioRxiv, 2020.01.21.914424. https://doi.org/10.1101/2020.01.21.914424
Popejoy, A. B., Domanska, D. E., & Thomas, J. H. (2020). BioRxiv, 2020.01.11.902759. https://doi.org/10.1101/2020.01.11.902759
Gichuki, D. K., Ma, L., Zhu, Z., Du, C., Li, Q., Hu, G., Zhong, Z., Li, H., Wang, Q., & Xin, H. (2019). . PeerJ, 7, e8201. https://doi.org/10.7717/peerj.8201
Publications are tagged with how they use, extend or reference Galaxy. This batch of pubs were tagged as:
468 : Methods 214 : UsePublic 81 : UseMain 78 : Workbench 54 : RefPublic 48 : UseLocal 26 : Tools 24 : Reproducibility 15 : IsGalaxy 6 : Cloud 5 : Shared 5 : Unknown 3 : Education 3 : HowTo 3 : Other 3 : Visualization 1 : Project
Other News
And so has the Galaxy ToolShed. And we are darn happy about it. Many thanks to Nicola Soranzo and Marius van den Beek for leading this years-long community wide effort.

The semi-annual update of the Galaxy Statistics Page happened. Stuff is up…

If you’re a UK-based researcher working from home, you might require some additional computing power. The Earlham Institute offers Galaxy and CyVerse UK cloud-based bioinformatics resources to help.

Read ScienceNode’s writeup on Galaxy and watch their interview with James Taylor at Gateways 2019.

Sema Elif Eski (ULB, BE) and Simon van Heeringen (Radboud Universiteit, NL) received the #UseGalaxy poster prizes at the Applied Bioinformatics in Life Sciences Conference in Leuven.





