July 2016 Galaxy News
Welcome to the July 2016 Galactic News, a summary of what is going on in the Galaxy community.
If you have anything to include in the next News, please send it to [Galaxy Outreach](mailto:outreach AT galaxyproject DOT org).
GCC2016 is done!
And it was again a smashing success! For the 5th year in a row we had over 200 participants signed up for GCC events, including hackathons, training, and the two day meeting. The meeting featured accepted presentations and keynotes, posters, the new Visualization Showcase and Software Demo sessions, lightning talks, and birds-of-a-feather meetups.
GCC2016 was held June 25-29 at Indiana University in Bloomington, Indiana, United States. This was the 7th annual gathering of the Galaxy community.
Slides for accepted and lightning talks are online, as are most posters. Remaining slides and video of training and talks will also be posted online this month.
Thanks to everyone for making GCC2016 a success, and looking forward to seeing you at GCC2017!
Upcoming Events
Several upcoming events were announced / highlighted at GCC2016:
GCC2017: 26-30 June 2017, Montpellier, France
GCC returns to Europe in 2017!
The 2017 Galaxy Community Conference (GCC2017) will be held 26-30 June in Montpellier, France. Montpellier is home to three universities, and is the agronomy hotspot in France. The city is home to half a million people is the third largest city of France’s Mediterranean coast.
GCC2017 will feature hackathons, two days of training and a two day meeting highlighting the best that data intensive biology has to offer.
All conference events will be at the Le Corum Conference Center, located in central Montpellier, next to the train station, and a 15 minute ride from the Montpellier International Airport. GCC2017 is hosted by the French Institute of Bioinformatics (IFB).
Swiss-German Galaxy Days
There will be a Swiss-German Galaxy Tour this fall featuring a range of events over 2 days in October. Events will held in Freiburg (Germany).
Registration is free, but space is limited on both days. We recommend you register soon to secure your spot for one or both days of the SG2016Tour: Register Now
Galaxy Admin Training, November 7-11, Salt Lake City, Utah
The first ever Galaxy Project Admin Training workshop will be held November 7-11 in Salt Lake City Utah. The workshop features a 2 day basics session followed by a 3 day advanced session. There is a separate registration for each session, and participants may sign up for one or both. We are working with the Utah High Performance Computing Center at the University of Utah for cluster access during the advanced session.
The introductory session will be at the Salt Lake City Library, and the advanced session will be on the University of Utah (the “U”) campus. Workshop housing is on the U as well.
The curriculum will be published and registration will open later this month. Watch this space. This workshop is the week before Supercomputing’16 meets in Salt Lake.
European Galaxy Developer Knowledge Exchange
ELIXIR, the European life-science Infrastructure for Biological Information, aims to orchestrate the collection, quality control, share-ability and archiving of large amounts of biological data produced by life science experiments and the resources to compute with this data.
Galaxy is widely used in the ELIXIR community. The French ELIXIR node (IFB) and the ELIXIR Galaxy Working Group will organize a knowledge exchange workshop in January 2017 (date tbc) in Strasbourg (France). This workshop will be for developers that want to set up a Galaxy instance, wrap tools and use Galaxy for data analysis and visualisation.
Galaxy AustralAsia Meeting (GAMe 2017), 3-9 February, Melbourne
The Galaxy Australasia Meeting (GAMe 2017) will be held 3-9 February in Melbourne Australia. The schedule has been set:
3 February
Using Galaxy Training
Morning and afternoon sessions covering 4 topics in two parallel tracks
4-5 February
Meeting
A two day meeting of the Galaxy AustralAsian communities featuring accepted and invited talks, posters and panel discussions on best practices, data intensive research, and the computational infrastructure to support it.
6-9 February
Admin Training
Four days of intense hands-on training on Galaxy Administration, based on the Galaxy Admin Training offered in the US in November.
Instructors and speakers will include experts from Australia and Asia, as well as members of the Galaxy Team. Training sessions and instructors will be announced shortly. Registration and abstract submission will open in August. Watch this space.
Other upcoming events
There are a plentitude of Galaxy related events coming up. Here’s what’s coming up over the next fe months:
| |
Designates a training event offered by GTN member(s) |
See the Galaxy Events Google Calendar for details on other events of interest to the community.
New Papers
82 new publications referencing, using, extending, and implementing Galaxy were added to the Galaxy CiteULike Group in June.
Some highlights from last month:
-
Integration and Visualization of Translational Medicine Data for Better Understanding of Human Diseases by Venkata Satagopam, Wei Gu, Serge Eifes et al. Big Data, Vol. 4, No. 2. (1 June 2016), pp. 97-108, doi:10.1089/big.2015.0057
-
biosigner: a new method for the discovery of significant molecular signatures from omics data by Philippe Rinaudo, Samia Boudah, Christophe Junot, Etienne A. Thevenot, Frontiers in Molecular Biosciences, Vol. 3, No. 26. (2016), doi:10.3389/fmolb.2016.00026
-
CART – a chemical annotation retrieval toolkit by Samy Deghou, Georg Zeller, Murat Iskar et al., Bioinformatics (02 June 2016), btw233, doi:10.1093/bioinformatics/btw233
-
Experiences in integrated data and research object publishing using GigaDB by Scott C Edmunds, Peter Li, Christopher I Hunter et al., In International Journal on Digital Libraries (2016), pp. 1-13, doi:10.1007/s00799-016-0174-6
Tagged Publications
The new papers were tagged with:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | Cloud | 1 | Other | 1 | Shared | 15 | UseMain | |||
| - | HowTo | - | Project | 3 | Tools | 14 | UsePublic | |||
| 4 | IsGalaxy | 6 | RefPublic | - | UseCloud | - | Visualization | |||
| 47 | Methods | 5 | Reproducibility | 6 | UseLocal | 22 | Workbench |
Who’s Hiring

The Galaxy is expanding! Please help it grow.
- PhD student or postdoctoral researcher, University of Freiburg, Freiburg, Germany.
- Postdoctoral Researcher: Forest Genomics Database and Software Developer, University of Connecticut. ”… data sharing among partner databases through Galaxy modules.” Deadline July 31.
- Software developer and Post-docs, Gehlenborg Lab, Harvard Medical School, Boston, Massachusetts, United States
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we’ll put it in the Galaxy News feed and include it in next month’s update.
New Public Galaxy Servers
One new publicly accessible Galaxy servers was added to the list in June:
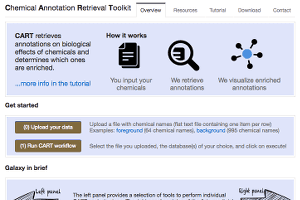
Chemical Annotation Retrieval Toolkit (CART)
- Link:
- Domain/Purpose:
- Retrieves annotations on biological effects of chemicals and determines which ones are enriched.
- Comments:
- From “CART – a chemical annotation retrieval toolkit” by Samy Deghou, Georg Zeller, Murat Iskar, Marja Driessen, Mercedes Castillo, Vera van Noort, and Peer Bork. Bioinformatics (2016), doi: 10.1093/bioinformatics/btw233 “it matches an input list of chemical names into a comprehensive reference space to assign unambiguous chemical identifiers. In this unified space, bioactivity annotations can be easily retrieved from databases covering a wide variety of chemical effects on biological systems. Subsequently, CART can determine annotations enriched in the input set of chemicals and display these in tabular format and interactive network visualizations, thereby facilitating integrative analysis of chemical bioactivity data.”
- Source code and an easy-to-install command line tool are also available.
- User Support:
- See the “Tutorial” and “Contact” tabs on the website.
- Quotas:
- Supports anonymous access and creation of user logins.
- Sponsor(s):
- Structural and Computational Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
New Semi-Public Galaxy Servers
And one new set of Semi-Public Galaxy server was added in June. Semi-public servers aren’t fully publicly accessible, but that are accessible to a larger group than just the host institution’s members.
Norway: NeLS
-
Links:
-
Eligibility:
- Life Science researchers from Norwegian academic institutions can log in with their FEIDE accounts (federation of home institution credentials). Other research collaborators can apply for a NeLS account.
-
Comments:
- The Norwegian e-Infrastructure for Life Sciences (NeLS) consists of a network of Galaxy servers at five Norwegian universities accessible via single sign-on.
- All the NeLS Galaxy servers are connected to the NeLS Portal which provides centralized storage and sharing capabilities.
- NeLS develops and maintains several analysis pipelines offered as Galaxy workflows.
- The NeLS infrastructure is operated by the Norwegian node of Elixir.
-
User Support:
- The Norwegian Bioinformatics Platform has a help desk that can be reached by emailing contact@bioinfo.no
-
Quotas:
- NeLS Galaxy instances have some temporary storage available, while the bulk volumes of data are managed through the NeLS centralized storage and the Norwegian national storage services.
-
Sponsor(s):
Galaxy Community Hubs
 |
 |
 |
| Share your training resources and experience now | Share your experience now |
One new community log board entry was added:
Releases
blend4php 0.1 alpha
We are pleased to announce the alpha release of the blend4php package, a PHP wrapper for the Galaxy API. It follows the lead of BioBlend which provides a Python package for interacting with Galaxy and CloudMan—hence the use of ‘blend’ in the name of this package. blend4php currently offers a partial implementation of the Galaxy API and includes support for datasets, data types, folder contents, folders, genomes, group roles, groups, group users, histories, history contents, jobs, libraries, library contents, requests, roles, search, tools, toolshed repositories, users, visualizations and workflows.
The motivation for development of this library is for integration with Tripal, an open-source toolkit for creation of online genomic, genetic and biological databases. Integration of Tripal and Galaxy will allow the community research databases to provide next-generation analytical tools to their users using Galaxy. However, this library was created independently of Tripal to support integration of any PHP application with Galaxy.
Please see the API documentation page for full information.
Planemo 0.27.0
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. Planemo 0.27.0 was released in June. Some highlights:
- Use ephemeris to handle syncing shed tools for workflow actions. 1c6cfbb
- More planemo testing enhancements for testing artifacts that aren’t Galaxy tools. Pull Request 491
- Implement
docker_galaxyengine type. eb039c0, Issue 15 - Enhance profiles to be Dockerized Galaxy-aware. Pull Request 488
- Add linter for DOI type citation - thanks to @mvdbeek. Pull Request 484
See the release history.
galaxy-lib 16.7.8 - 16.7.9
galaxy-lib is a subset of the Galaxy core code base designed to be used as a library. This subset has minimal dependencies and should be Python 3 compatible. It’s available from GitHub and PyPi.
Highlights of June releases include:
- Updates for recent changes to Galaxy and cwltool.
- Updates to include Galaxy library for verifying test outputs and the latest dev changes to Galaxy.
Earlier Releases
Galaxy v16.04
The Galaxy Committers team is pleased to announce the April 2016 (v16.04) release of Galaxy.
Please note that Python 2.6 is no longer supported as of this release. See the 16.04 release announcement for details.
Highlights
Tool Profile Versions
Tools may now declare which version of Galaxy they require. Tools requiring 16.04 or newer will have new default behaviors (such as using exit code for error detection) that should simplify tool development. See PR #1688.
Embedded Pulsar Job Runner
Galaxy can now start a Pulsar application embedded within the Galaxy process itself. This allows using Pulsar’s job staging and isolation without requiring a RESTful web service or a message queue. This is enabling usegalaxy.org to run jobs to on the new JetStream cloud. See PR #2057.
New Chemical Datatypes
Galaxy now detects and supports many molecular datatypes. See PR 1941. Thanks to Björn Grüning (@bgruening).
| Github New % git clone -b master https://github.com/galaxyproject/galaxy.gitUpdate to latest stable release % git checkout master && git pull --ff-only origin masterUpdate to exact version % git checkout v16.04 |
BitBucket Upgrade % hg pull% hg update latest_16.04 |
|
| See the Get Galaxy page for additional details regarding the source code locations. | ||
Galaxy Docker Image 16.04
And, thanks to Björn Grüning, there is also now a Docker image for Galaxy 16.04 as well.
CloudMan 16.04
We just released an update to Galaxy CloudMan on AWS. CloudMan offers an easy way to get a personal and completely functional instance of Galaxy in the cloud in just a few minutes, without any manual configuration or imposed quotas. Once running, you have complete control over Galaxy, including the ability to install new tools.
This is a minor update release with the following changes:
- Galaxy 16.04 update
- Availability on Amazon’s Ireland region
- A couple of bug fixes
See the CHANGELOG for a more complete set of changes.
Pulsar 0.7.0
Pulsar 0.7 was released in April. Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
And the rest …
Other Galaxy packages that haven’t had a release in the past four months can be found on GitHub.
ToolShed Contributions
Did not get there this month. Look for a double edition next month.
Other News
- biojs2galaxy: Continuous integration of BioJS components into Galaxy A fast tool to convert BioJS components into Galaxy visualization plugins. Warning: This tool is in a ALPHA stage. Use at your own risk.
- v1.0.0 release of ASaiM framework, an open-source Galaxy-based opinionated framework dedicated to microbiota sequence analyses